Novel deep learning method enables clinic-ready automated screening for diabetes-related eye disease
Deep-learning in hospitals
In recent years, clinics have taken first steps towards artificial intelligence and deep learning to automate medical screenings. However, training a deep learning algorithm for accurate screening and diagnosis prediction requires large sets of annotated data and clinics often struggle with expensive expert labelling.
Researchers were therefore looking for ways to reduce the need for costly annotated data while still maintaining the high performance of the algorithm.
Use case diabetic retinopathy
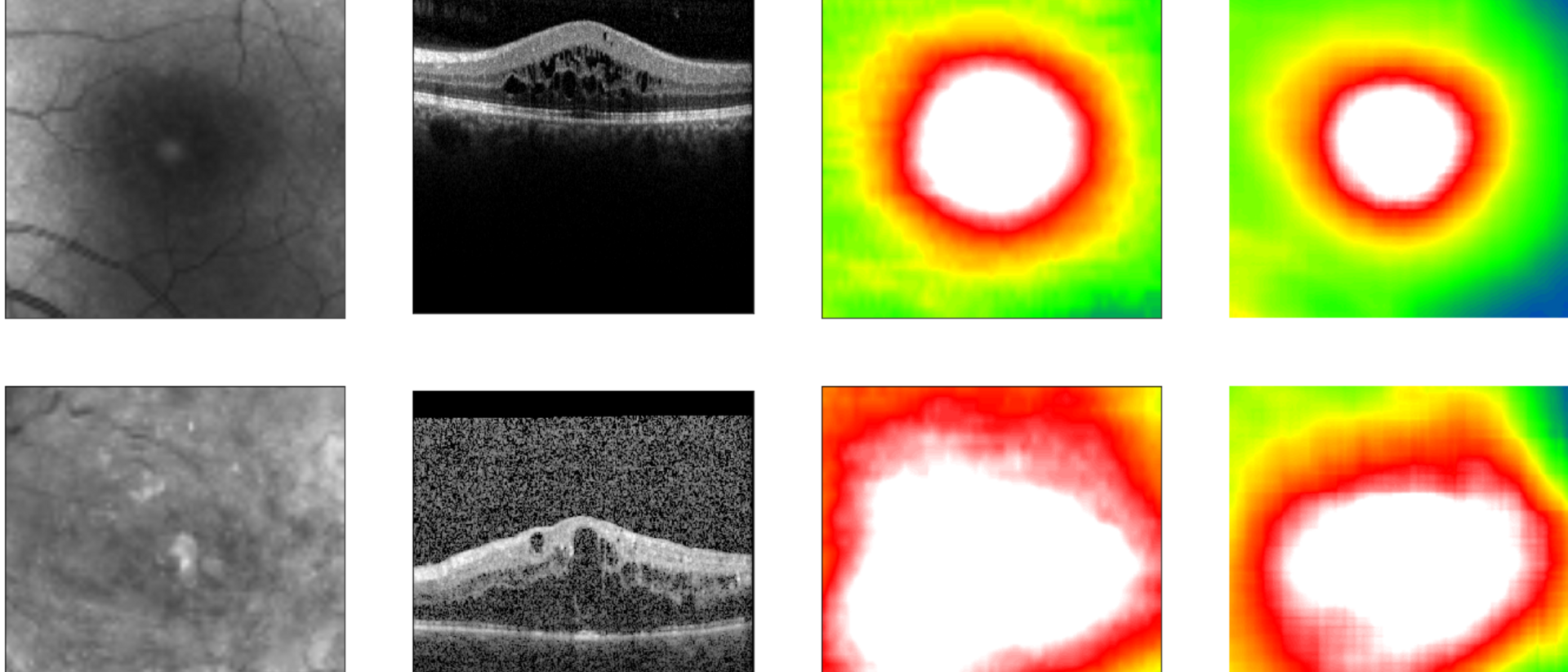
Diabetic retinopathy is a diabetes-related eye disease damaging the retina and can ultimately lead to blindness. Measuring the retinal thickness is an important procedure to diagnose the disease in risk patients. To do so, most clinics take photographs of the fundus – the surface of the back of the eye. In order to automate the screening of these images, clinics started to apply deep learning algorithms. These algorithms require large sets of fundus images with expensive annotations in order to be trained to screen correctly.
The LMU University Eye Hospital Munich owns a population-size data set containing over 120,000 unannotated fundus and co-registered OCT images. OCT (optical coherence tomography) allows for precise information about the retinal thickness but is not commonly available in every eye care center. The LMU provided their data to researchers from the TUM and Helmholtz Zentrum München pioneering in the field of artificial intelligence in health.
Pre-training under “self-supervision”
“Our goal was to use this uniquely large set of fundus and OCT images to develop a method which will reduce the need of expensive annotated data for algorithm training”, says Olle Holmberg, first author of the study from the TUM School of Life Sciences.
The group of researchers developed a novel method called “cross modal self-supervised retinal thickness prediction” and applied it to pre-train a deep learning algorithm with the LMU data set. In this use case, cross modal self-supervised learning allowed the algorithm to teach itself to recognize unannotated fundus images with different OCT-derived retinal thickness profiles, predicting the thickness information directly from the fundus. By accurately predicting retinal thickness, a key diagnostic feature for diabetic retinopathy, the algorithm was then able to learn how to predict screening outcomes.
High performance with a quarter of training data
This novel method shrunk the need for expensive annotated data to train the deep learning algorithm significantly. When applied in automated screenings for diabetes retinopathy, it achieved the same diagnostic performance, both, compared to previous algorithms which had required much more training data and compared to human experts.
“We reduced the need for annotated data by 75 percent”, states Fabian Theis, Professor of Mathematical Modelling of Biological Systems at the TUM and Director of the Institute of Computational Biology at Helmholtz Zentrum München. “Sparse annotated data is a grand challenge in medicine. It is one of our goals to develop methods that work with less data and that can then potentially be applied in many settings. Our use case in diabetic retinopathy is ready for immediate use in clinics and is a perfect example of how AI can improve the daily business of clinics and thus everybody’s health.”
Moreover, an additional reduction in size, meaning number of parameters, was achieved in the algorithm itself. The novel method enables up to 200 times smaller algorithms. This could be a crucial benefit to deploying them on mobile and embedded devices which is also important in clinical settings.
Applications beyond diabetic retinopathy
Beyond diabetic retinopathy, the novel method allows for further clinical applications where much unannotated data is available but expert annotations are scarce, such as age-related macular degeneration (AMD).
Holmberg et al., 2020: Self-supervised retinal thickness prediction enables deep learning from unlabeled data to boost classification of diabetic retinopathy. Nature Machine Intelligence, DOI: 10.1038/s42256-020-00247-1
The self-supervised pre-trained algorithm from this study is available on https://github.com/theislab/DeepRT.
Technical University of Munich
Corporate Communications Center
- V. Schulz / K. Baumeister
- presse@tum.de
- Teamwebsite
Contacts to this article:
Prof. Dr. Fabian Theis
Technical University of Munich
Chair of Mathematical Modeling of Biological Systems
Boltzmannstr. 3, 85748 Garching
Phone: +49 (89) 289 - 18386
e-Mail: theis@mytum.de